The dataset we will be using for this tutorial comes from the transcriptomics world, a paper published in BMC Genomics in 2013 entitled "Transcriptomic characterization of cold acclimation in larval zebrafish" (Long et al., 2013).
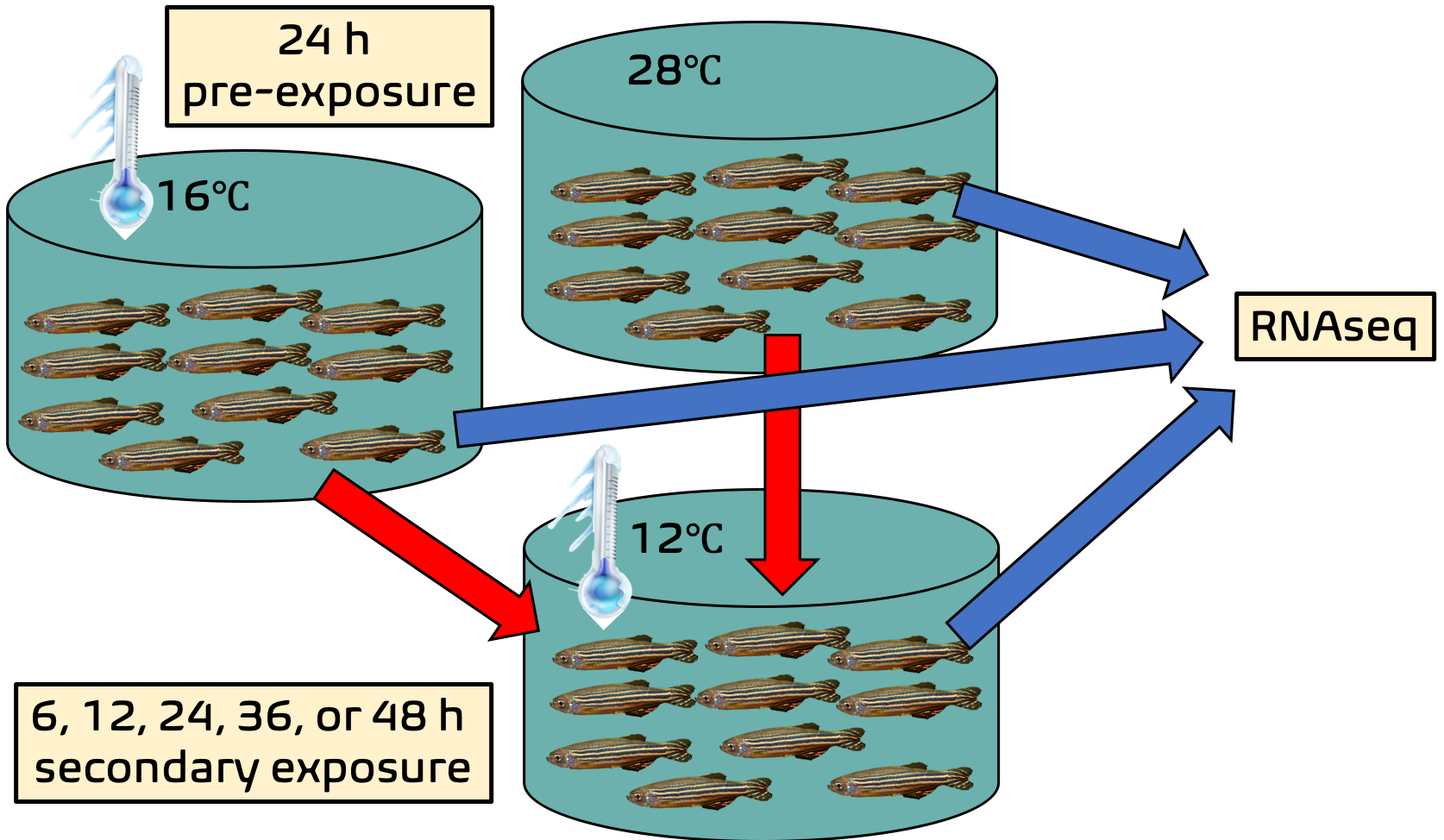
The authors pre-exposed 96 zebrafish larvae to cold stress for 24 hours, which for these zebrafish is defined as 16 degrees Celsius. They then upped the ante (after removing some samples for sequencing) to extreme cold stress, wherein the zebrafish larvae were exposed to 12 degrees Celsius temperatures. They exposed the larvae to the super-cold temperature for 6, 12, 24, 36, or 48 hours before also sending these samples for RNAseq analysis.
The authors found that genes responsible for RNA splicing, ribosome biogenesis, and protein catabolism were most substantially affected by cold exprosure. They also found that the cold-stressed larvae switched out genes via alternative splicing and promoter switching , which allowed them to use slightly different forms of genes or promoters to induce the transcription process that would work a little more smoothly in the wake of their new, icy world.
The figure below briefly summarizes the basic idea of the experiment (imagine the fish pictures are larvae):
- Long, Y., Song, G., Yan, J., He, X., Li, Q., & Cui, Z. (2013). Transcriptomic characterization of cold acclimation in larval zebrafish. BMC Genomics, 14(1), 612.