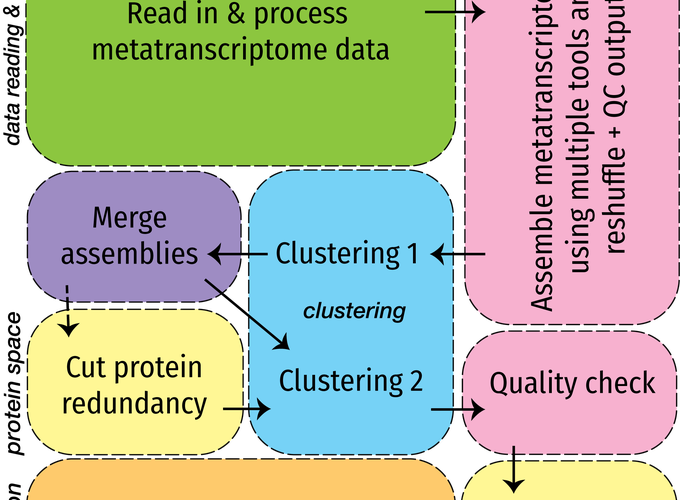
Metatranscriptomics is a useful probe into the members of an ecological community, capturing community members and their potential functions in a single sequencing effort. Because transcribed RNA is sequenced, via thorough processing of metatrancriptomes, we can decipher both the members constituting a community as well as the genes that each member was expressing at the instant that the sample was collected. Metatranscriptomics is a relatively immature pursuit as compared to genomics or transcriptomics. However, even more palpable is the lack of thorough analysis and readily available tools for metatranscriptomes from the environment. More complicated still is the use of metatranscriptomes for eukaryotes from the environment, many of which have not been previously or thoroughly sequenced and have complex genetic features. Eukrhythmic is an approach to consolidating existing assembly effort for environmental eukaryotes. The pipeline includes assembly via multiple external assembly tools, and then a process of consolidating those assemblies based on sequential clustering steps. Also included with the pipeline is the option to perform downstream functional and taxonomic annotation of retrieved transcripts.
The goal of this project is both to validate and provide support for the efforts of [marine] microbial ecologists that use metatranscriptomics to assess community structure and function, and to provide a tool that researchers can use to assemble their own environmental metatranscriptomes. Some questions associated with this project are: 1. Can we recover complex communities from shuffled metatranscriptomic sequencing reads? 2. Does the choice of assembler affect the quality of the assembly output?
Project Repository: https://github.com/AlexanderLabWHOI/eukrhythmic