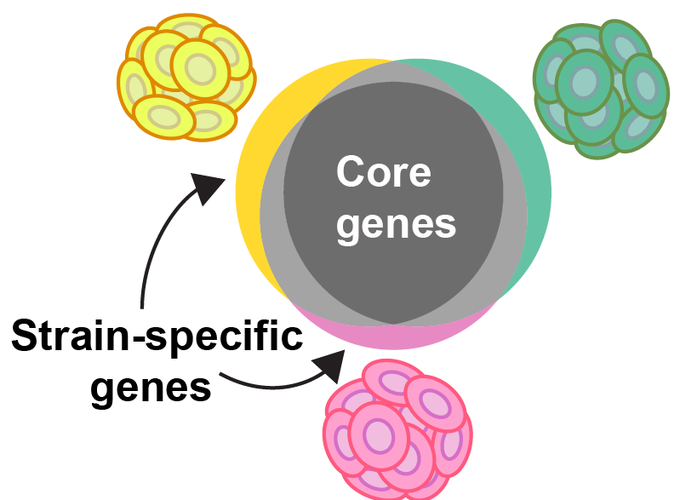
Coccolithophores, a group of calcifying marine phytoplankton within the phylum Haptophyta, play a significant role in marine biogeochemical cycles, particularly in those of carbon and sulfur. Emiliania huxleyi is one of the most abundant extant coccolithophore species in the ocean, typified by its cosmopolitan distribution and ability to form large blooms. Intra-specific variability in physiology (e.g. enzymatic rates, morphology, growth rates) of E. huxleyi has long been known in cultured isolates. The capacity for this variability was revealed through the genome sequencing of many E. huxleyi strains, the results of which suggested that E. huxleyi has a ‘pan genome,’ with gene content varying across strains. Intensive study of another ecologically significant taxa with a pan genome, the cyanobacteria Prochlorococcus, has connected the variability of the pan genome to its niche separation and biogeographical distribution. Similar to Prochlorococcus, intra-specific genomic variability may underpin the success of E. huxleyi across diverse environmental conditions and underlie its observed physiological variability. Here we are combining a series of genomic, computational field surveys, and laboratory-based experiments designed to identify the function of the pan genome in E. huxleyi ecology and biogeography. Through this project, we will address the questions:
- Is there structure to the variable gene content across strains in the pan genome of E. huxleyi?
- Does the environment select for particular strains of E. huxleyi based on the content of the variable portion of the pan genome?
- Does the composition of the variable gene set determine strain success under shifting environmental conditions?
Funding for this project provided by the National Science Foundation